Form of reactions:
Form of reactions:
Mostly 2 n
Occasionally 1 + heat n
Occasionally 1 + heat n
Basic phenomenon:
Basic phenomenon:
Representation of all possible reactions, independent of molecular location and any exogenous factors:
In[]:=
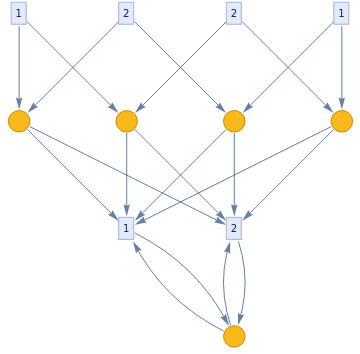
ResourceFunction["TokenEventGraph"][{{1,2}->{3,4}},{1,1,2,2},2,"TokenDeduplication"->False]
Out[]=
Implicitly assuming “well stirred” [AKA “equilibrium dynamics”]
Things to add: e.g. location of the molecules. Are they in different compartments? Or different positions
Microscopically molecules can have different energies [energies are roughly determined by temperature]
Ultimately a molecule has position (x) and momentum (p).... [Usually the internal degrees of freedom will be distributed according to temperature; unless e.g. one excites them with a nonthermal photon]
Biology is not well stirred...
Reactions without enzymes: mostly small molecules
Are the enzymes free floating or bound to a membrane [typically partially embedded]
[ proteins, but sometimes with extra attachments ]
[ proteins, but sometimes with extra attachments ]
Most collisions in a “chemical process” are elastic... [the elastic collisions knit together the structure of the fluid]
Molecular orientation determines whether the reaction will happen
Inelastic collisions: vibrational/rotational/etc. excitation + actual reaction
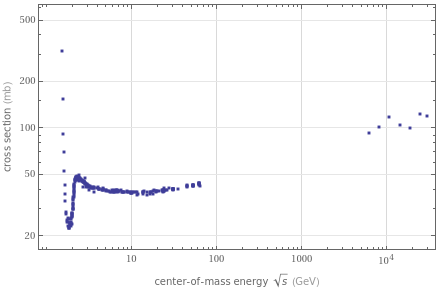
n σ v
In[]:=
Out[]=
In[]:=
UnitConvert[Quantity[40,"Millibarns"]]//N
Out[]=
In[]:=
Sqrt[%]
Out[]=
In[]:=
ResourceFunction["TokenEventGraph"][{{1,2}->{1,2}},{1,1,2,2},2,"TokenDeduplication"->False,"EventDeduplication"->False]
Out[]=
In[]:=
(*Functiontogenerateacausalgraphfromthecollisionslist.*)collisionsToCausalGraph[collisions_]:=Module[{particles,instants,edges},(*Extractparticlesfromlistofcollisions*)particles=Union@@collisions[[All,;;2]];(*Generatecausalgraphfromcollisions.Constructadirectededgewheneveranuninterruptedsequenceoftwocollisionsinvolvesthesameparticle.*)edges=Catenate[Function[particle,instants=SplitBy[Select[collisions,#[[1]]===particle||#[[2]]===particle&],Last];Catenate[BlockMap[DirectedEdge@@@Tuples[#]&,instants,2,1]]]/@particles];(*Finally,constructthegraph*)Graph[DeleteDuplicates@edges,Sequence[GraphLayout->"LayeredDigraphEmbedding",AspectRatio->1/GoldenRatio,ImageSize->Large]]]
In[]:=
(*Runasimulationandextractthecausalgraph*)With[{num=15,steps=50,boxSize=5,dimension=2,rad=1,stepSize=0.1},Module[{collisions,initialPos,initialVel},(*Randominitialconditions*)initialPos=RandomReal[{-#,#}&@(boxSize-rad),{num,dimension}];initialVel=RandomPoint[Ball[ConstantArray[0,dimension]],num];(*Runasimulation.Ignorefirst20%toeliminatetransientroleofinitialconditions.*)collisions=Select[ResourceFunction["HardSphereSimulation"][initialPos,initialVel,Sequence[N[boxSize],N[stepSize],N[rad],steps]][[2]],#[[3]]>Round[steps/5]&];(*Generatecausalgraph*)collisionsToCausalGraph[collisions]]]
Out[]=
Analogy: every hyperedge is a molecule
We can create atoms of space, but in chemistry we usually don’t (except e.g. from a solvent)
Rate of collision vs. rate of reaction
Rate of collision vs. rate of reaction
Substitution reaction:
[[ Which atom goes to which molecule in the reaction? ]] [ usually you can tell ]
Different levels of description
Different levels of description
Consider 22 string rewriting.....
First approximation: conserve the number of each character
This shows the ensemble of all possible events for these particular inputs
(Bug for not not deduplicating the initial state)
Levels:
Levels:
1. All possible reactions between individual molecules
2. A particular sequence of reactions between individual molecules
3. Sequence of reaction between molecular species [typical chemistry level]
Level 1 provides probabilities for level 2; level 2 gives “concentrations” for level 3 [ in level 3 we just count the molecules; we don’t give each one a name ]
Level 1 : QM ; level 2 : SM ; level 3 : thermochemistry
“Activity” vs “concentration”
“Activity” vs “concentration”
A + B C + D E + F
A1 + B1 C1 + D1
C1 + D678 -> E1 + F1
C1 + D678 -> E1 + F1
With catalysts / enzymes ... might not work this way...
Solvay process: