Spontaneous Oscillations in Yeast Chemostat Cultures
Spontaneous Oscillations in Yeast Chemostat Cultures
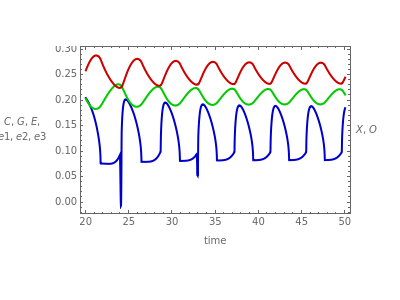
There have been numerous reports of spontaneous oscillations occurring in chemostat cultures of yeast Saccharomyces cerevisiae. A cybernetic model of yeast metabolism has been used to predict the appearance and disappearance of these sustained oscillations in response to changes in the chemostat operating parameters (dilution rate (1/hour), oxygen transfer coefficient a (1/hour), inlet glucose concentration (g/l), and inlet ethanol concentration (g/l)). This Demonstration allows the user to investigate how bioreactor concentrations of cell mass (gdw/l), dissolved oxygen (mg/l), glucose (g/l), ethanol (g/l), intracellular amounts of the carbohydrate mass fraction (g/g cdw), and three hypothesized enzymes are predicted to respond to changes in the operating parameters.
D
k
L
G
in
E
in
X
O
G
E
C
The buttons correspond to bioreactor concentrations that vary with time, while the sliders correspond to operating parameters that are independent of time. The different colors enable the user to determine which curve corresponds to which of the selected variables.
Details
Details
Large values of maximum time should be used with care because due to the relative complexity of the equations being solved, lengthy computation times may result.
Taken from K. D. Jones and D. S. Kompala, "Cybernetic Model of the Growth Dynamics of Saccharomyces Cerevisiae in Batch and Continuous Cultures," Journal of Biotechnology, 71(1), 1999 pp. 105–131.
Permanent Citation
Permanent Citation
David J. W. Simpson, Dhinakar S. Kompala
"Spontaneous Oscillations in Yeast Chemostat Cultures"
http://demonstrations.wolfram.com/SpontaneousOscillationsInYeastChemostatCultures/
Wolfram Demonstrations Project
Published: March 14, 2008